“Why can’t we write the entire twenty-four volumes of the Encyclopedia Britannica on the head of a pin?”
Richard Feynman offered up this daunting challenge (with a rather paltry $1000 prize) at his famous 1959 Caltech lecture “There’s Plenty of Room at the Bottom” – a seminal event in the history of nanotechnology. In 1985, Tom Newman, then a Stanford grad student, completed the challenge. Newman reduced the first paragraph of A Tale of Two Cities to the necessary 1/25,000 of its size using a focused beam of electrons to carve out the lettering.


Since then, nanoscale writing has become something of a scientific competition (who can make it smaller) as well as an interesting marker of progress in nanotechnology since Feynman’s vision over 50 years ago. In 1989, a team at IBM precisely arranged 35 xenon atoms to spell out the company logo. The current record holders are a Stanford team, who in 2009, manipulated subatomic pieces on the order of 0.3 nanometers to spell out the university’s initials. A scanning tunneling microscope was used to position single carbon monoxide molecules on a sliver of copper. Electrons in the copper bounce around, spreading out as waves and rebounding off the carefully placed carbon monoxide molecules. The specific interference patterns of these electron waves encode the letters. Hari Manoharan, author of the paper, put this accomplishment in perspective, “One bit per atom is no longer the limit for information density. There’s a grand new horizon below that, in the subatomic regime. Indeed, there’s even more room at the bottom than we ever imagined.”
Yet, Feynman’s challenge, writing out an entire book, remains unmet. The techniques previously described are laborious, costly, and above all painfully slow. All of these techniques are based on manipulating single atoms one at a time. The recent field of DNA nanotechnology offers a possible solution.
Most people are surprised to learn that DNA makes for an effective building material at the nanoscale; yet, nucleic acids are life’s information storage molecule of choice. The key to building small is encoding the assembly information into the molecules themselves rather than using external forces to arrange them. The main advantage of DNA is that the sequence of nucleotides in a strand can be precisely controlled and pairing rules are well understood. Moreover, strands of DNA with complementary sites will always spontaneously assemble into the most favorable structure.

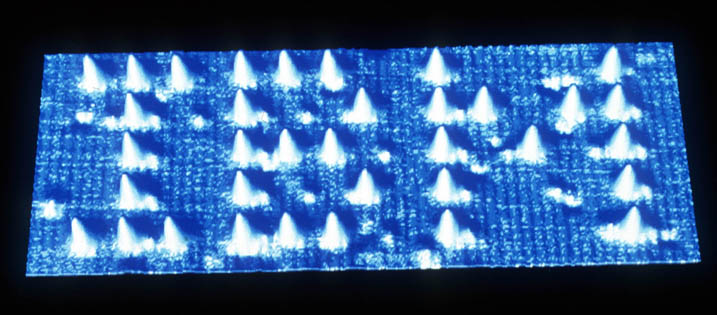
In a 2006 Nature article, Paul Rothemund coined the fanciful term “DNA Origami” to describe his successful manipulation of DNA strands into a variety of shapes. His revolutionary technique utilized a single long strand of genomic DNA from a bacteriophage, which was coiled, twisted, and stacked by small custom “staple” strands which bind to complementary sites. By winding the long strand back and forth with staple strands, Rothemund folded six different 2-D shapes including squares, triangles, and stars – demonstrating that this technique was applicable to any shape within a 100nm diameter.
Rothemund’s most significant achievement was developing a method to pattern the 2-D sheets. He designed staple strands that would stick up from the flat lattice, increasing the height of the nanostructure at desired locations. A world map as well as the word “DNA” were successfully created and visualized. Perhaps the most surprising part of the method is its simplicity – staple strands are mixed with the long strands, heated for 2 hours and viola! The staple strands can be designed with computer algorithms and synthesized at relatively cheap cost, which makes scaling up easy. How appropriate that one day humanity’s knowledge may be recorded in the same molecules that encode our lives.

One response to “Reading Between the Atoms – Writing on the Nanoscale”
Thanks on your marvelous posting! I genuinely enjoyed reading it, you can be a great author.I will be sure to bookmark your blog
and may come back at some point. I want to encourage one
to continue your great work, have a nice morning!