Gram Negative Bacteria and Antibiotic Resistance
Highly resistant to most antibiotics, intricately-structured Gram-negative bacteria (GNB), such as E. coli and K. pneumoniae are the cause of some of the world’s most rampant infections. These bacteria continuously develop powerful antibiotic resistance mechanisms, making common antibiotics increasingly ineffective.1
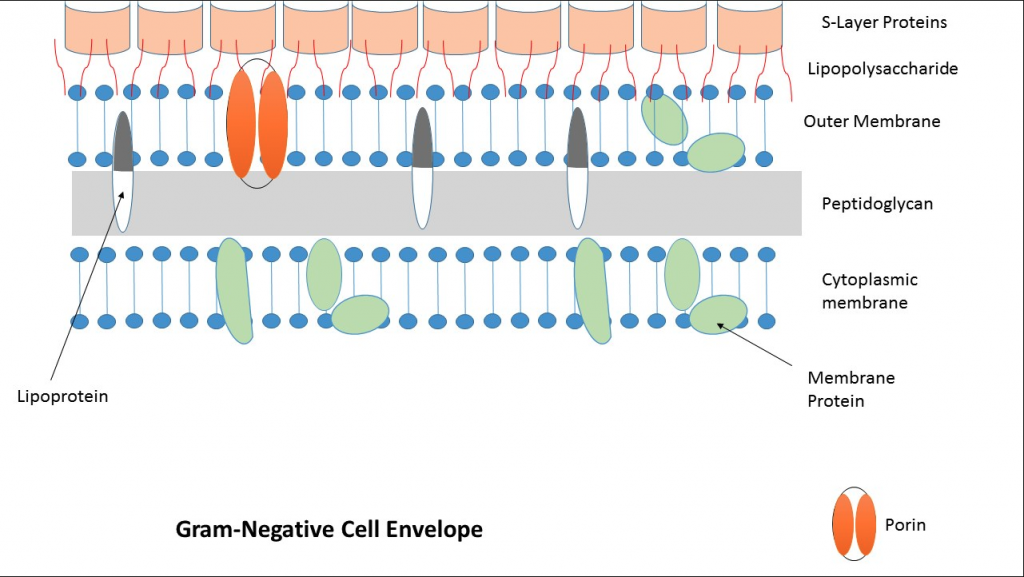
Most antibiotics work by disrupting processes essential to bacterial growth. In order for them to be effective, they need to be able to reach the interior of the bacterial cell. However, GNB combats this with an additional external layer, called the outer membrane. The outer membrane is an extremely effective defense against antibiotics because it tightly controls the types of molecules permitted to pass into the cell. In addition to this innate selectivity, mutations in membrane transporters cause general incompatibility with antibiotics, which furthers antibiotic impermeability. A common example is in a channel-forming transport protein called a porin, which moves nutrients and antibiotics into the cell. A porin is susceptible to loss of function mutations which can make passive diffusion, a method of cross-membrane transportation, extremely difficult. And while GNB have other efficient methods to intake nutrients, these mutations can make it incredibly difficult for antibiotics to pass through the outer membrane. Due to the advantageous nature of this mutation for GNB, it becomes inherited by offspring, allowing for continued bacterial growth in the presence of antibiotics, providing a form of inherent antibiotic resistance.2,3
Introducing Efflux Inhibitors (EPIs)
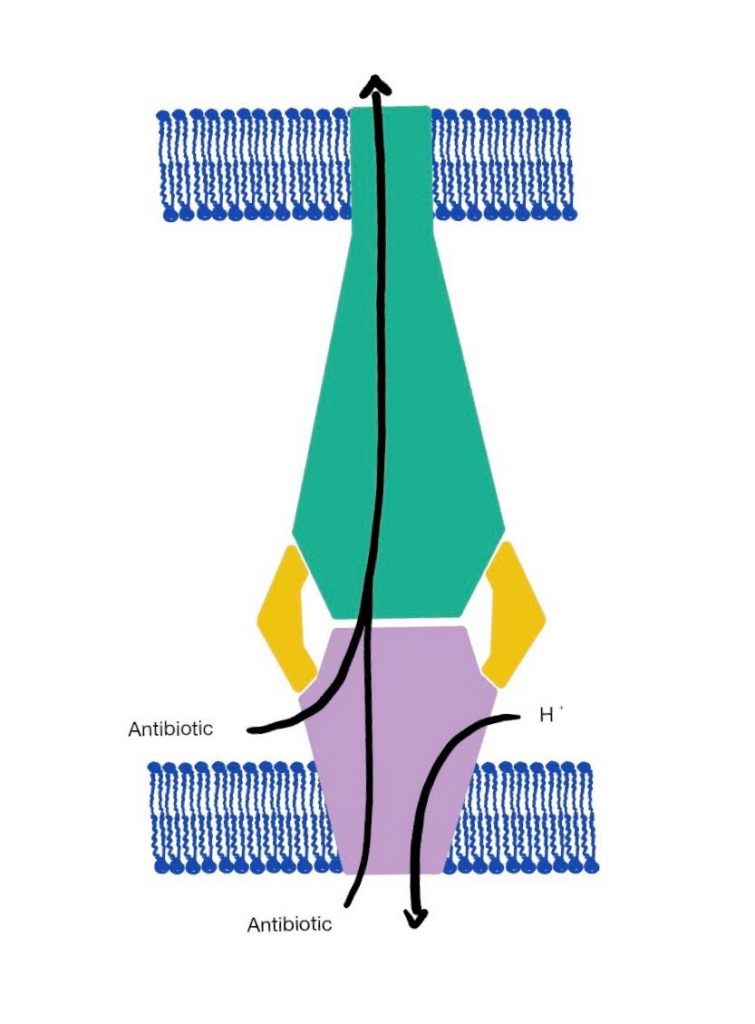
For the past decade, scientists have been exploring efflux pumps as a way to exploit the defensive system of GNB. Efflux Pumps are protein transporter complexes that have the ability to pump antibiotics back out of the cell. In addition to the outer membrane, they serve as one of GNB’s secondary modes of defense against antibiotics. Efflux Pumps rest against the cytoplasmic membrane and force antibiotics out by actively transporting them back through the outer membrane using special channels that connect to the outside of the cell, as seen in Figure 2. When unmodified, Efflux pumps can render an antibiotic ineffective, preventing it from reaching its intracellular target.
Multi-Drug Efflux Pumps, or efflux pumps that have the ability to recognize a wide range of antibiotics, are often key contributors to widespread antibiotic resistance.5 A substantial amount of research has been conducted on methods to counteract the resistance caused by Multi-Drug Efflux Pumps, most notably surrounding the use of Efflux Pump Inhibitors (EPIs).
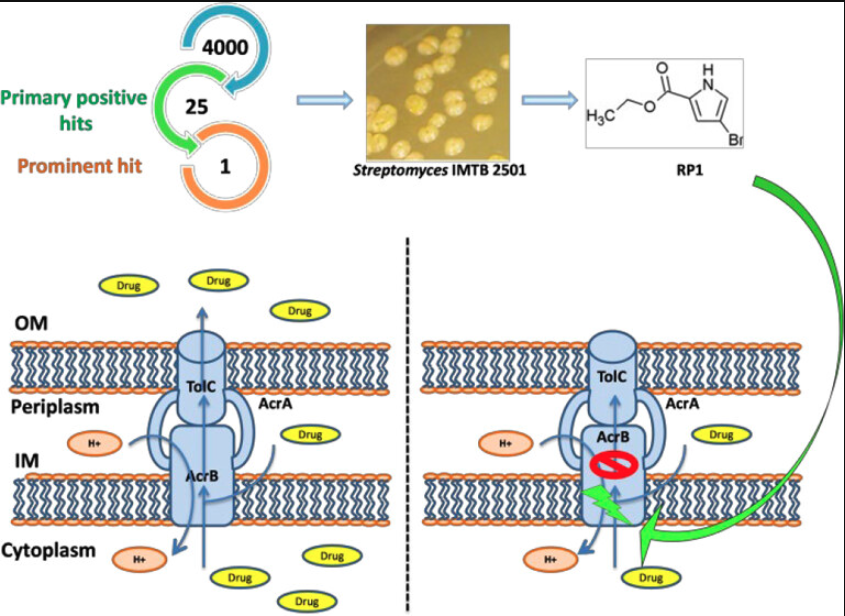
Although there are numerous methods an EPI can impede efflux activity, there are two commonly employed mechanisms. The first involves adding substrates to the cell to provide competitive inhibition for the antibiotics against the efflux pumps. In this case, EPIs outcompete antibiotics to be captured by the Efflux Pump. This gives antibiotics a higher likelihood of not being transported out of the cell. The second involves “stealing” the protons, ions, or ATP needed for the active transportation through the efflux pump. This impairs the efflux pump, as energy from the protons, ions, or ATP is needed for its active transport processes. An example of this can be seen with the AcrAB-TolC Efflux Complex, one of the strongest antibiotic efflux pumps in E. coli. An EPI known as RP1 provides competitive inhibition for the antibiotics within the cell. RP1 functions by binding to efflux receptors within the ArcB portion of the efflux pump. RP1 outcompetes the antibiotics for these binding spots, allowing the antibiotic to remain in the cell while RP1 is pumped out instead.
Discovering EPI candidates
A common way of discovering potential EPI targets is by performing a gene knockout, where scientists disrupt the genetic code to inactivate an efflux pump and then observe what has changed within the cell.This can be seen in an experiment related to resistance in M. abscessus, which is an aggressive species of bacteria distantly related to M. Tuberculosis, one of the world’s deadliest diseases. 2 genes in M. abscessus were found to be involved in resistance to Ethionamide, an antibiotic treatment for MultiDrug Resistant Tuberculosis (MDR-TB). In their experiment, researchers found that by knocking out the two target genes, the bacteria became susceptible to Ethionamide. This data, along with more supporting statistics, led the team to believe that genes may encode for an efflux pump with the ability to pump Ethionamide out of the cell. This discovery can then be used to experiment with possible EPI candidates, which can help make Ethionamide a potent antibiotic against MDR-TB and other Non-Tuberculosis Mycobacterium.11
Limitations
Although scientists have made large advancements with EPIs, there are still a few limitations with its usage. Unfortunately, Multi-Drug Efflux Pumps have the ability to recognize many EPIs, and can therefore pump them out of cells as well. In addition to this, many potential EPIs are toxic, and therefore can’t be used clinically. While these downsides are still being researched, several EPIs have proven to be highly successful, giving many previously ineffective antibiotics another opportunity to reach their target.12
This science and the future
Our current knowledge of both efflux pumps and the application of EPIs only reflects a fraction of their importance in GNB. Scientists are actively studying the presence, purpose, and activity of many efflux pumps in GNB. With the rate of research today, new potential EPI candidates in are constantly being researched in order to develop antibiotics that are more effective against GNB.12 As these antibiotic resistance persists, it is vital that scientists continue to counteract this harmful trend by furthering our understanding of both efflux pumps and EPIs. The hope is that these scientific endeavors can eventually produce a straightforward, resistance-free solution to the rampant, disease-causing GNB worldwide.
Acknowledgements
I’d like to thank Anna Christenson, a Molecular and Cellular Biology Ph.D. student at University of California, Berkeley, and Anaya Sánchez, a Ph.D Candidate at University of California, Berkeley, for so generously reviewing my article and providing plenty of detailed and thoughtful feedback.
References
- Oliveira, J., & Reygaert, W. C. (2023, August 8). Gram-negative bacteria – statpearls – NCBI bookshelf. https://www.ncbi.nlm.nih.gov/books/NBK538213/
- Delcour AH. Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta. 2009 May;1794(5):808-16. doi: 10.1016/j.bbapap.2008.11.005. Epub 2008 Nov 27. PMID: 19100346; PMCID: PMC2696358.
- Breijyeh, Z., Jubeh, B., & Karaman, R. (2020). Resistance of Gram-Negative Bacteria to Current Antibacterial Agents and Approaches to Resolve It. Molecules, 25(6), 1340. https://doi.org/10.3390/molecules25061340
- Allgood, S. C., Su, C.-C., Crooks, A. L., Meyer, C. T., Zhou, B., Betterton, M. D., Barbachyn, M. R., Yu, E. W., & Detweiler, C. S. (2023). Bacterial Efflux Pump Modulators Prevent Bacterial Growth in Macrophages and Under Broth Conditions that Mimic the Host Environment (p. 2023.09.20.558466). bioRxiv. https://doi.org/10.1101/2023.09.20.558466
- A Microbe-Derived Efflux Pump Inhibitor of the Resistance-Nodulation-Cell Division Protein Restores Antibiotic Susceptibility in Escherichia coli and Pseudomonas aeruginosa. (n.d.). https://doi.org/10.1021/acsinfecdis.1c00281
- Farhana, A., & Khan, Y. S. (2023, April 17). Biochemistry, lipopolysaccharide – StatPearls – NCBI Bookshelf. https://www.ncbi.nlm.nih.gov/books/NBK554414/
- Nishino, K., Yamasaki, S., Nakashima, R., Zwama, M., & Hayashi-Nishino, M. (2021). Function and Inhibitory Mechanisms of Multidrug Efflux Pumps. Frontiers in Microbiology, 12. https://www.frontiersin.org/articles/10.3389/fmicb.2021.737288
- Sharma, A., Gupta, V. K., & Pathania, R. (2019). Efflux pump inhibitors for bacterial pathogens: From bench to bedside. The Indian Journal of Medical Research, 149(2), 129–145. https://doi.org/10.4103/ijmr.IJMR_2079_17
- Tambat, R., Mahey, N., Chandal, N., Verma, D. K., Jangra, M., Thakur, K. G., & Nandanwar, H. (2022). A microbe-derived efflux pump inhibitor of the resistance-nodulation-cell division protein restores antibiotic susceptibility in escherichia coli and pseudomonas aeruginosa. ACS Infectious Diseases, 8(2), 255–270. https://doi.org/10.1021/acsinfecdis.1c00281
- Pagès, J.-M., & Amaral, L. (2009). Mechanisms of drug efflux and strategies to combat them: Challenging the efflux pump of Gram-negative bacteria. Biochimica Et Biophysica Acta, 1794(5), 826–833. https://doi.org/10.1016/j.bbapap.2008.12.011
- Rodriguez, R., Campbell-Kruger, N., Camba, J. G., Berude, J., Fetterman, R., & Stanley, S. (2022). MarR-Dependent Transcriptional Regulation of mmpSL5 induces Ethionamide Resistance in Mycobacterium abscessus (p. 2022.10.03.510743). bioRxiv. https://doi.org/10.1101/2022.10.03.510743
- Abdel-Karim, S. A.-A., El-Ganiny, A. M., El-Sayed, M. A., & Abbas, H. A. (2022). Promising FDA-approved drugs with efflux pump inhibitory activities against clinical isolates of Staphylococcus aureus. PLOS ONE, 17(7). https://doi.org/10.1371/journal.pone.0272417
- Gil, T., Laborda Martínez, P., Ochoa Sánchez, L., Martínez, J., & Hernando-Amado, S. (2023). Efflux in Gram-negative bacteria: What are the latest opportunities for drug discovery? Expert Opinion on Drug Discovery, 18, 1–16. https://doi.org/10.1080/17460441.2023.2213886
- Li, X.-Z., Plésiat, P., & Nikaido, H. (2015). The Challenge of Efflux-Mediated Antibiotic Resistance in Gram-Negative Bacteria. Clinical Microbiology Reviews, 28(2), 337–418. https://doi.org/10.1128/cmr.00117-14
- (PDF) Efflux in Gram-negative bacteria: What are the latest opportunities for drug discovery? (n.d.). Retrieved October 2, 2023, from https://www.researchgate.net/publication/370871220_Efflux_in_Gram-negative_bacteria_what_are_the_latest_opportunities_for_drug_discovery
- Vaara, M. (1992). Agents that increase the permeability of the outer membrane. Microbiological Reviews, 56(3), 395–411.
- Zhang, Z. (2022). Hacking the Permeability Barrier of Gram-Negative Bacteria. ACS Central Science, 8(8), 1043–1046. https://doi.org/10.1021/acscentsci.2c00750
Image References
Figure 1: Patel, H. (2016, June 23). Gram Negative Cell Wall. Wikimedia Commons. https://commons.wikimedia.org/wiki/File:Gram-Negative_Cell_Envelope.jpg
Figure 3: Tambat, R., Mahey, N., Chandal, N., Verma, D. K., Jangra, M., Thakur, K. G., & Nandanwar, H. (2022a). A microbe-derived efflux pump inhibitor of the resistance-nodulation-cell division protein restores antibiotic susceptibility in escherichia coli and pseudomonas aeruginosa. ACS Infectious Diseases, 8(2), 255–270. https://doi.org/10.1021/acsinfecdis.1c00281